- MGB Personalized Medicine Biobank Genomics Core
MGB Personalized Medicine Biobank Genomics Core
At Your Service
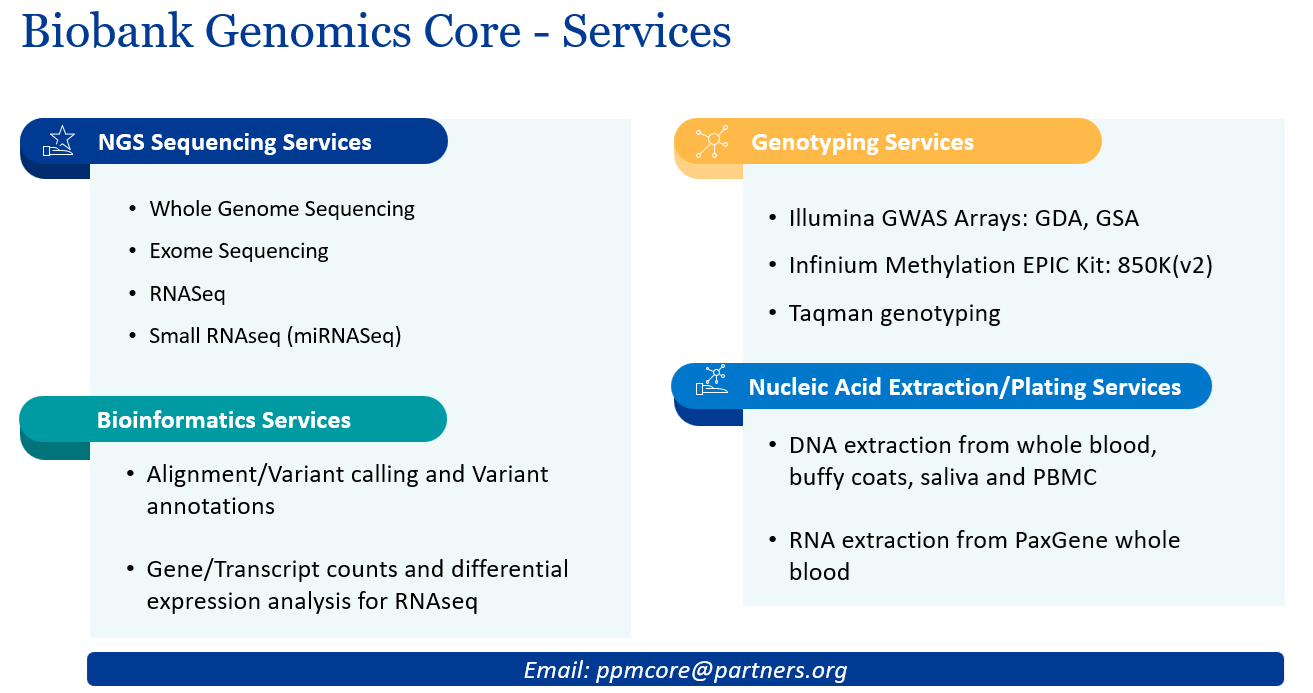
At Mass General Brigham Personalized Medicine Biobank Genomics Core (BGC), we offer genotyping and next generation sequencing services to support your project. With over a decade of experience and a proven track record of helping MGB investigators achieve their research aims, our goal is to make sequencing/genotyping technologies accessible to the research community.
Our services are listed below, please contact us for additional information on any of our services, or for pricing. We also provide support documentation (letter of support, workflow description, quotes) to support grant applications.
Accounts and Ordering
All projects are managed through our internal Laboratory Information Management System (StarLIMS), with orders for services submitted via PCMS (Core Management System).
Submit an Order/Service Request
Please submit a service request prior to submitting samples. To do this, you must first set up a PCMS account. You also must have a valid MGB IRB # and a Grant number or PO number. Detailed instructions can be found in this user guide.
Where is MGB Personalized Medicine Biobank Genomics Core located?
MGB Personalized Medicine
65 Landsdowne St., 3rd floor
Cambridge, MA 02139-4232
Directions
How do I receive my data?
You will be contacted when data is available. Depending on size, data can be shipped to you on an external hard drive. We can also coordinate other delivery mechanisms. Data will remain available from MGB Personalized Medicine for 3 months.
Contact MGB PM Biobank Genomics Core
For Illumina Next-Gen Sequencing and Genotyping quotes and customer inquiries, please email PPMCore@partners.org
For DNA/RNA Extraction and sample handling service quotes and customer inquiries, please email PPMCENTRALBIOBANK@partners.org
For help placing an order or other IT-related inquiries, please email PMBGCIT@mgb.org